Simple example of decoding: the Haxby data¶
Here is a simple example of decoding, reproducing the Haxby 2001 study on a face vs cat discrimination task in a mask of the ventral stream.
Script output:
Downloading data from http://data.pymvpa.org/datasets/haxby2001/MD5SUMS ...
...done. (0 seconds, 0 min)
Downloading data from http://data.pymvpa.org/datasets/haxby2001/subj1-2010.01.14.tar.gz ...
...done. (912 seconds, 15 min)
Extracting data from /home/mfpgt/nilearn_data/haxby2001/e55ccad24e48bcce63c6a2ea0eb83b59/subj1-2010.01.14.tar.gz...
...done.
First subject anatomical nifti image (3D) is at: /home/mfpgt/nilearn_data/haxby2001/subj1/anat.nii.gz
First subject functional nifti images (4D) are at: /home/mfpgt/nilearn_data/haxby2001/subj1/bold.nii.gz
[0.72727272727272729, 0.46511627906976744, 0.72093023255813948, 0.58139534883720934, 0.7441860465116279]
Python source code: plot_haxby_simple.py
### Load haxby dataset ########################################################
from nilearn import datasets
haxby_dataset = datasets.fetch_haxby()
# print basic information on the dataset
print('First subject anatomical nifti image (3D) is at: %s' %
haxby_dataset.anat[0])
print('First subject functional nifti images (4D) are at: %s' %
haxby_dataset.func[0]) # 4D data
### Load Target labels ########################################################
import numpy as np
# Load target information as string and give a numerical identifier to each
labels = np.recfromcsv(haxby_dataset.session_target[0], delimiter=" ")
# scikit-learn >= 0.14 supports text labels. You can replace this line by:
# target = labels['labels']
_, target = np.unique(labels['labels'], return_inverse=True)
### Keep only data corresponding to faces or cats #############################
condition_mask = np.logical_or(labels['labels'] == b'face',
labels['labels'] == b'cat')
target = target[condition_mask]
### Prepare the data: apply the mask ##########################################
from nilearn.input_data import NiftiMasker
mask_filename = haxby_dataset.mask_vt[0]
# For decoding, standardizing is often very important
nifti_masker = NiftiMasker(mask_img=mask_filename, standardize=True)
func_filename = haxby_dataset.func[0]
# We give the nifti_masker a filename and retrieve a 2D array ready
# for machine learning with scikit-learn
fmri_masked = nifti_masker.fit_transform(func_filename)
# Restrict the classification to the face vs cat discrimination
fmri_masked = fmri_masked[condition_mask]
### Prediction ################################################################
# Here we use a Support Vector Classification, with a linear kernel
from sklearn.svm import SVC
svc = SVC(kernel='linear')
# And we run it
svc.fit(fmri_masked, target)
prediction = svc.predict(fmri_masked)
### Cross-validation ##########################################################
from sklearn.cross_validation import KFold
cv = KFold(n=len(fmri_masked), n_folds=5)
cv_scores = []
for train, test in cv:
svc.fit(fmri_masked[train], target[train])
prediction = svc.predict(fmri_masked[test])
cv_scores.append(np.sum(prediction == target[test])
/ float(np.size(target[test])))
print(cv_scores)
### Unmasking #################################################################
# Retrieve the SVC discriminating weights
coef_ = svc.coef_
# Reverse masking thanks to the Nifti Masker
coef_img = nifti_masker.inverse_transform(coef_)
# Save the coefficients as a Nifti image
coef_img.to_filename('haxby_svc_weights.nii')
### Visualization #############################################################
import matplotlib.pyplot as plt
from nilearn.image.image import mean_img
from nilearn.plotting import plot_roi, plot_stat_map
mean_epi = mean_img(func_filename)
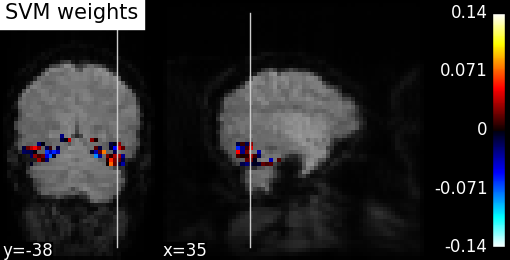
plot_stat_map(coef_img, mean_epi, title="SVM weights", display_mode="yx")
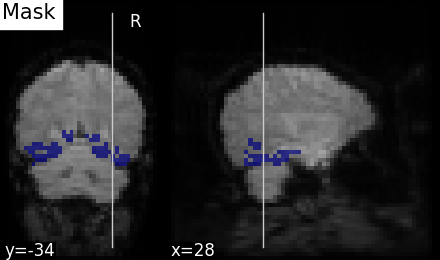
plot_roi(nifti_masker.mask_img_, mean_epi, title="Mask", display_mode="yx")
plt.show()
Total running time of the example: 926.93 seconds ( 15 minutes 26.93 seconds)